Resources

Our lab mainly utilizes massively parallel MD simulation package OpenMM using graphical processing units (GPUs). Our lab creates a high-performance supercomputing ecosystem equipped with ~150 GPUs (mostly GeForce 30 series + 32 RTX 4090 !), which enables a single simulation across multiple GPUs, or alternatively run a different simulation on each one at the same time.
GPUs/CPUs cluster
| Node | GPUs | CPUs | RAM | TFlops (FP32) |
|---|---|---|---|---|
| 15 | NVIDIA GeForce RTX 4090 x 10 | Intel(R) Xeon(R) Gold 6242R CPU x 40cores | 250GB | 825.8 |
| 18 | NVIDIA GeForce RTX 4090 x 6 | Intel(R) Xeon(R) Gold 6348 CPU x 112cores | 500GB | 495.48 |
| 19 | NVIDIA GeForce RTX 4090 x 6 | Intel(R) Xeon(R) Gold 6348 CPU x 112cores | 500GB | 495.48 |
| 20 | NVIDIA GeForce RTX 4090 x 10 | Intel(R) Xeon(R) Platinum 8360Y CPU x 72cores | 500GB | 825.8 |
| 1 | NVIDIA GeForce RTX 3090 x 8 | Intel(R) Xeon(R) Gold 6230 CPU x 32cores | 125GB | 284.64 |
| 3 | NVIDIA GeForce RTX 3090 x 10 | Intel(R) Xeon(R) CPU E5-2680 v4 x 28cores | 250GB | 355.8 |
| 11 | NVIDIA GeForce RTX 3090 x 12 | Intel(R) Xeon(R) Gold 6242R CPU x 40cores | 250GB | 426.96 |
| 12 | NVIDIA GeForce RTX 3090 x 12 | Intel(R) Xeon(R) Gold 6242R CPU x 40cores | 250GB | 426.96 |
| 13 | NVIDIA GeForce RTX 3090 x 8 | Intel(R) Xeon(R) Gold 6230R CPU x 52cores | 125GB | 284.64 |
| 16 | NVIDIA GeForce RTX 3090 x 10 | Intel(R) Xeon(R) Gold 6242R CPU x 40cores | 250GB | 355.8 |
| 5 | NVIDIA GeForce RTX 3080Ti x 8 | Intel(R) Xeon(R) Gold 6242R CPU x 40cores | 250GB | 272.8 |
| 6 | NVIDIA GeForce RTX 3080Ti x 12 | Intel(R) Xeon(R) CPU E5-2699 v4 x 44cores | 250GB | 409.2 |
| 14 | NVIDIA GeForce RTX 3080Ti x 12 | Intel(R) Xeon(R) Gold 6242R CPU x 40cores | 250GB | 409.2 |
| 17 | NVIDIA GeForce RTX 3080Ti x 12 | Intel(R) Xeon(R) Gold 6242R CPU x 48cores | 250GB | 409.2 |
The theoretical performance of our cluster is estimated to be 6,280 TFlops (note that the entire KISTI-5 Supercomputer Nurion’s theoretical performance is 25,700 Tflops!). These faciltiies turbocharge our research by significantly boosting our simulation speed. For more, see the benchmark (below).
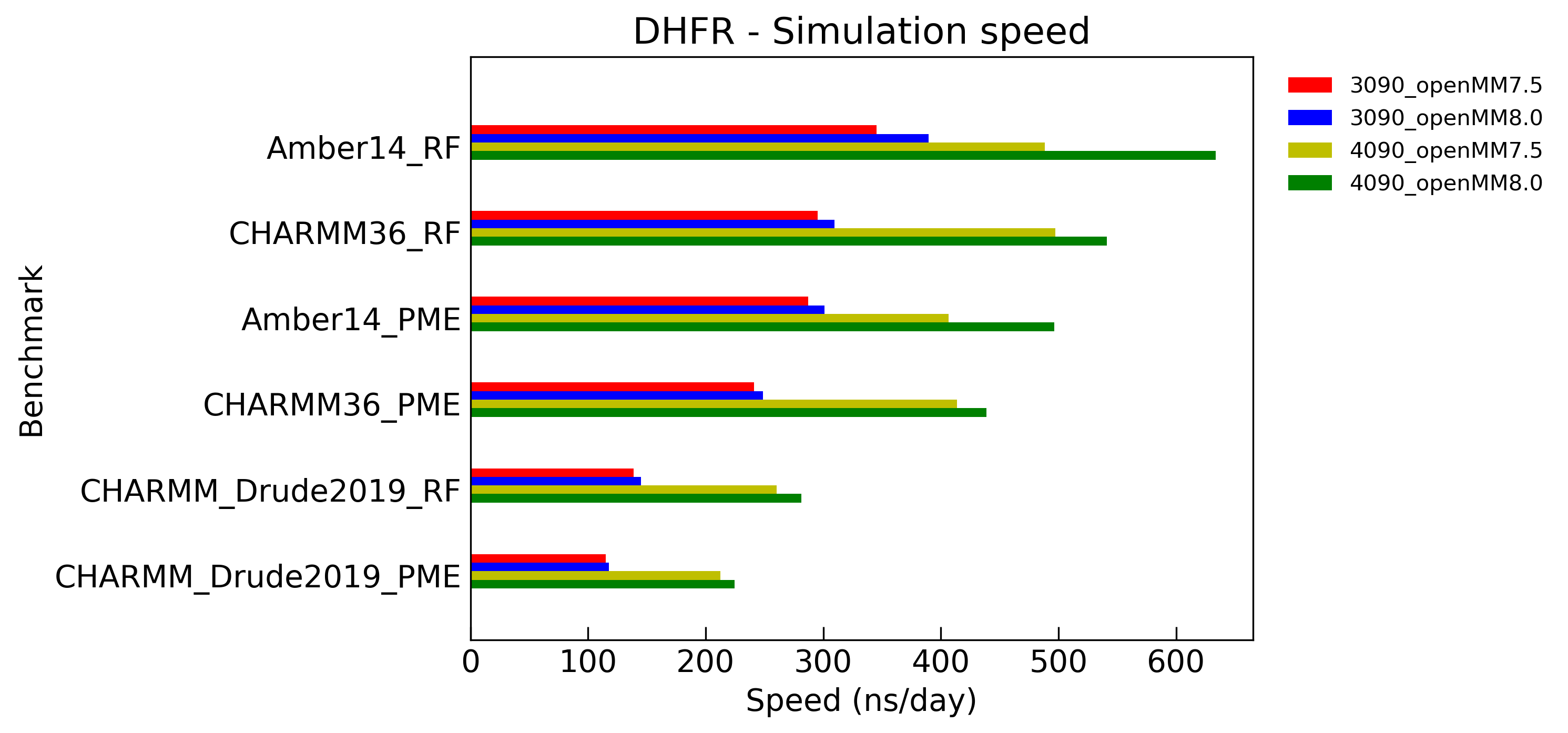
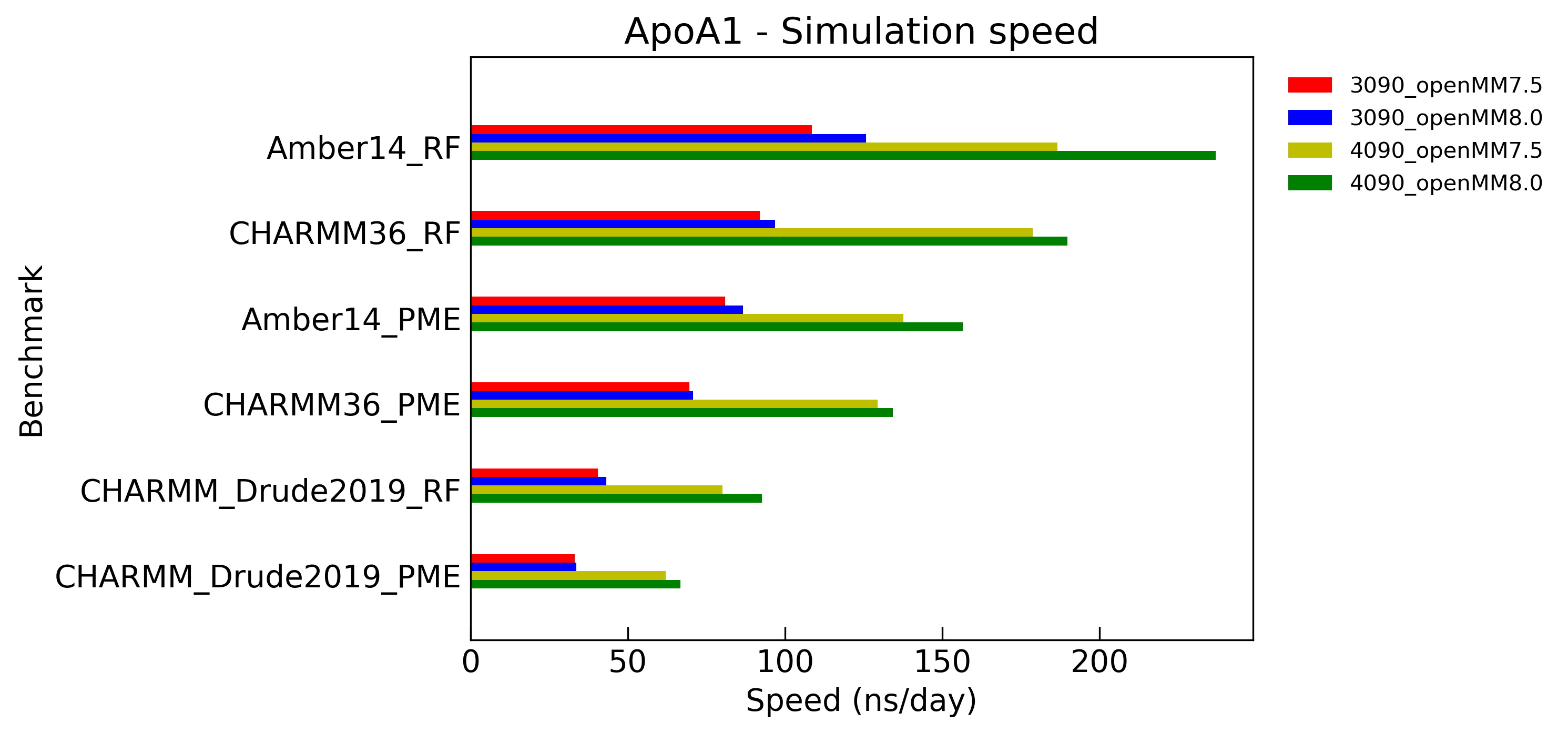
Simulation speed
Benchmarks of simulation speed on typical biochemical systems. Solvents are treated either explicit-RF or explicit-PME (1.2nm for cutoff distance). Langevin / Drude Langevin integrators are employed with a step size of 1 fs. The length of bonds involving hydrogen were constrained, and water molecules were fully rigid. CUDA platform on NVIDIA GeForce RTX 3090/4090 GPUs are used with mixed precision.
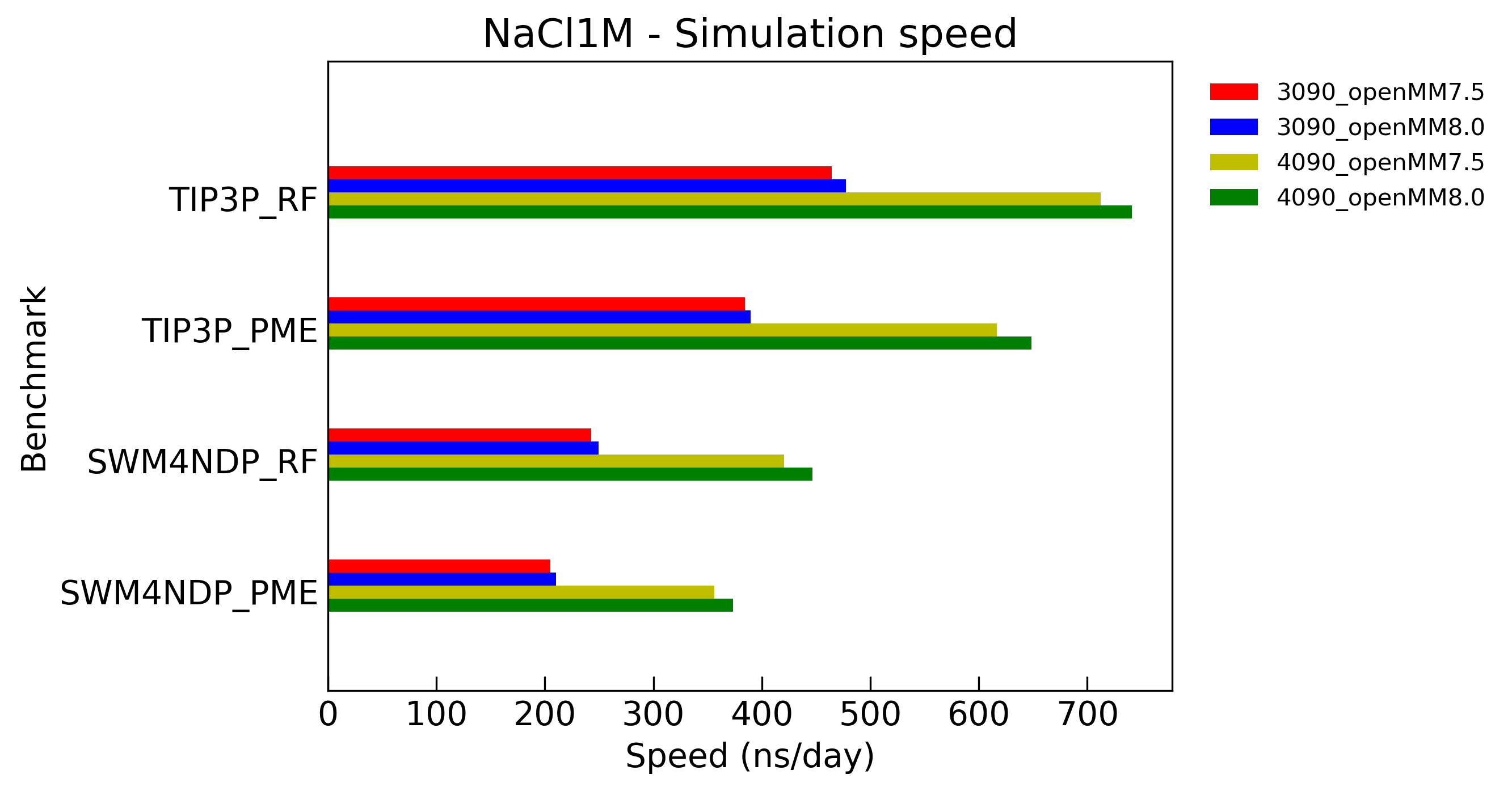
System: NaCl 1M Solution
5nm box, 3512 water molecules, 75 pair of Na+/Cl- ions, total 10686 atoms
Model: TIP3P, SWM4-NDP
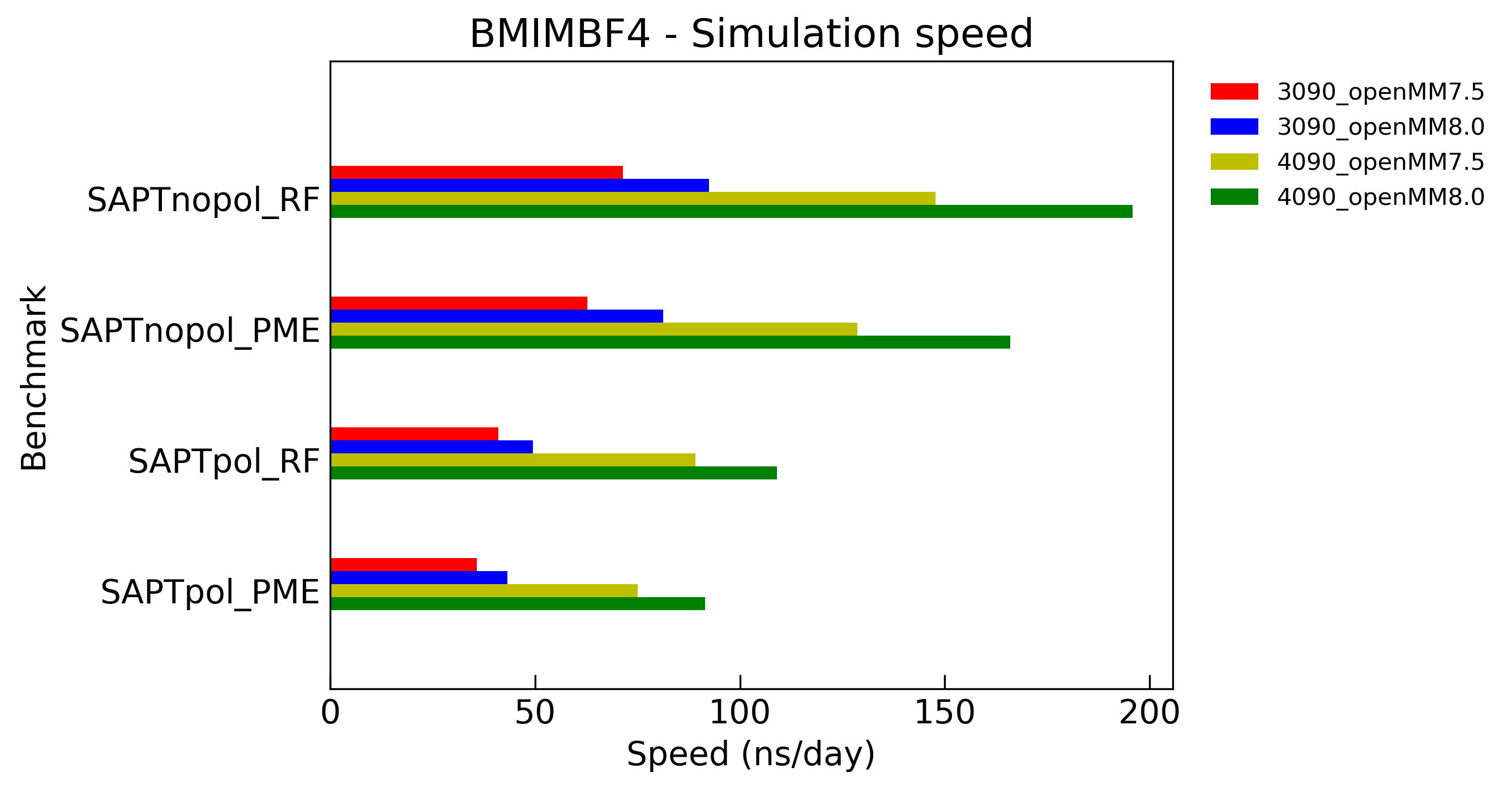
System: Ionic liquid BMIM+/BF4-
8.0nm box, 1600 pair of BMIM+/BF4- ions, total 48000 atoms
Model: SAPT, SAPT_Drude